GLOBAL Lipidomics Assay
At Li Node, we offer a state-of-the-art GLOBAL (Untargeted) Lipidomics Profiling service, utilizing both positive and negative ionization techniques for comprehensive lipidome analysis. Our cutting-edge method is capable of detecting, identifying, and relatively quantifying a vast array of lipids – over 5,000 in positive ionization and more than 2,000 in negative ionization.
Key Features of Our Service:
- High-Throughput Identification: Using MS/MS analysis, we typically identify approximately 1,000 lipids, while an additional 3,000 to 5,000 lipids are putatively identified through accurate mass-match.
- Deuterated Internal Lipid Standards: We employ deuterated lipid standards from 15 different lipid classes for data normalization. This approach ensures accurate, relative quantification of the lipids, enhancing the reliability of comparisons between samples.
- Comprehensive Service Package: Our service encompasses all aspects of lipidomics research – from sample preparation, lipid extraction, and LC-MS analysis to data analysis and lipid identification. We also include data normalization and a detailed statistical analysis, featuring PCA, PLS-DA, and Volcano plots.
- Results and Data Delivery: Clients receive a thorough summary of our analysis and statistical results. The detected lipids, their putative identifications (either by MS/MS spectral match or accurate mass match), and relative quantification data are neatly summarized in an Excel file. This file is not only comprehensive but also versatile, ready for further analysis such as uploading to MetaboAnalyst for advanced statistical or pathway analysis.

At Li Node, our lipidomics analysis services employ a meticulous and tiered approach for metabolite identification. This strategy ensures high accuracy and comprehensiveness in identifying a wide range of lipids. Here’s how our identification tiers are structured:
Tier 1 Identification:
- Criteria for Identification: Lipids in this tier are identified based on two key parameters:
- Mass error must be less than or equal to 5.0 mDa.
- MS/MS match score must be 500 or higher when compared to published databases.
- Range of Identification: Typically, we are able to identify between 150 and 900 lipids in this tier.
Tier 2 Identification:
- Enhanced Identification Criteria: This tier requires fulfilling three stringent criteria for lipid identification:
- Mass error ≤5.0 mDa.
- MS/MS match score ≥500.
- Isotope pattern match score (mSigma) ≤100, compared to published databases.
- Identification Capacity: In Tier 2, we typically identify between 100 and 500 lipids.
Tier 3 Identification:
- Putative Identification Based on MS Data: This tier involves putative identification, which is primarily based on mass-to-charge ratio (m/z) data matched to database hits.
- Extensive Identification Scope: In this tier, we can typically identify 3000 or more lipids.
Each tier in our metabolite identification process is designed to ensure the most accurate and comprehensive lipid profiling. By employing such a detailed and tiered approach, we can cater to the varying needs of lipidomic research, providing our clients with reliable and extensive data for their studies. Whether you are exploring a small set of specific lipids or conducting a broad survey of the lipidome, our service is equipped to deliver high-quality, detailed lipidomic analysis.
Academic/Government Clients:
- Base Price: $215 CAD per sample for projects with 20 samples or more.
- Small Batch Surcharge: For projects with 19 samples or less, an additional charge of $400 CAD is applied.
- Lipid Identification Fee: A flat fee of $635 CAD is added per project for lipid identification services.
- Sample Handling Fee: Each project also incurs a flat sample handling fee of $200 CAD.
Industry Clients:
- Base Price and Surcharges: The same base price and small batch surcharge as for academic/government clients apply.
- Overhead: An additional 30% overhead is added to the total cost for industry clients.
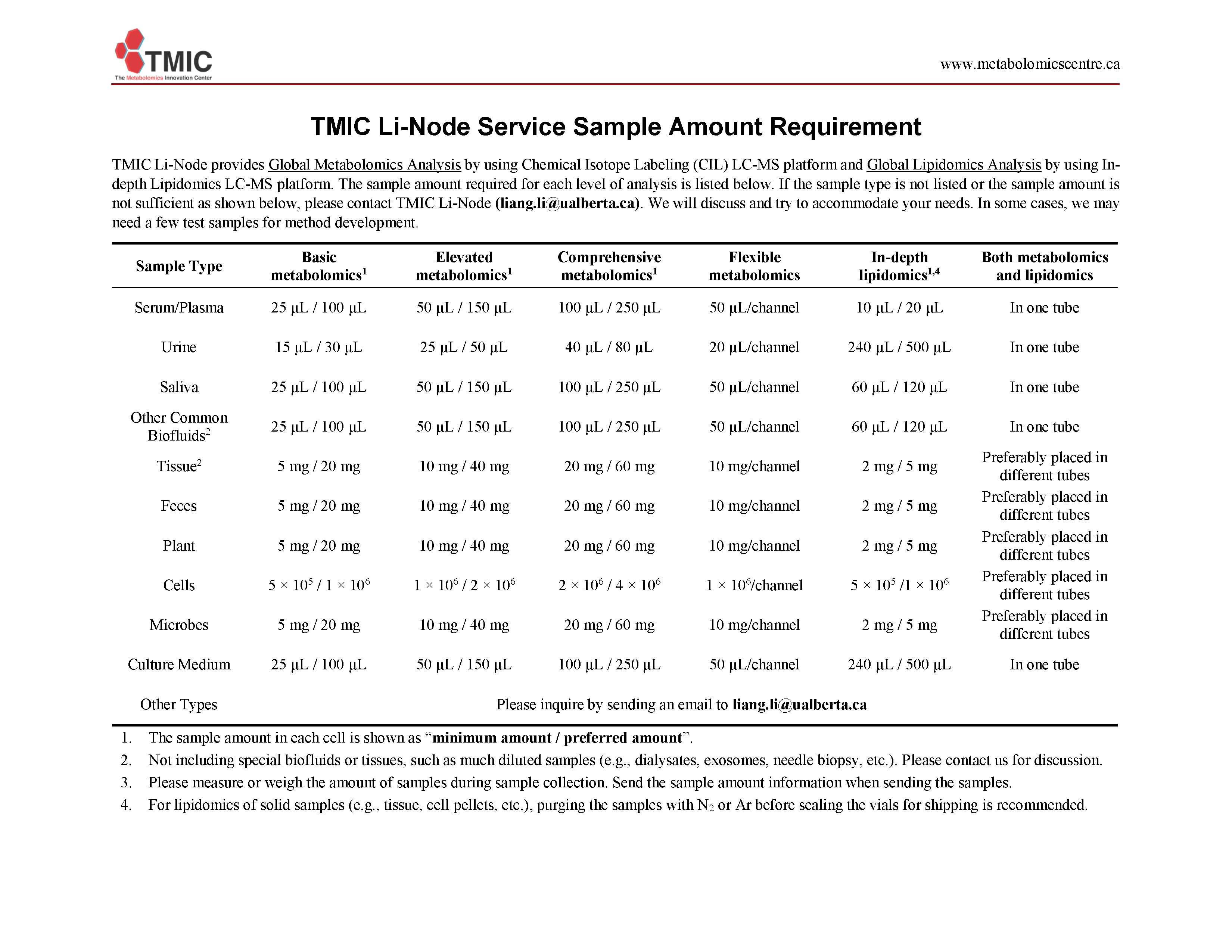
The sample amount required for each level of analysis is listed below. If the sample type is not listed or the sample amount is not sufficient as shown below, please contact TMIC Li-Node (liang.li@ualberta.ca). We will discuss and try to accommodate your needs. In some cases, we may need a few test samples for method development.
Should you require further information or have any inquiries, please visit our Contact Us page.










