GLOBAL Metabolomics Assay
Untargeted
To increase metabolome coverage and achieve accurate quantification for all detectable metabolites, we use a Chemical Isotope Labeling (CIL) Metabolomics Platform (Zhao et al., 2019; 2020). Our approach analyzes four submetabolomes: amine/phenol, carboxyl, carbonyl, and hydroxyl, covering 85%-95% of the metabolome. Users can choose to analyze one or more submetabolomes based on budget and desired coverage.
With one submetabolome analysis, we accurately quantify up to 2,500 metabolites in samples like urine, blood, plant extract, and cell extracts. Using all four analyses, we cover 7,000 to 10,000 metabolites per sample.
We handle sample preparation, metabolite extraction, chemical labeling, LC-MS analysis, data analysis, metabolite ID, and statistical analysis (PCA, PLS-DA, Volcano plots). A summary of the analysis and results, including peak pairs, metabolite IDs, and relative quantification data, will be provided in an Excel file for further analysis, such as uploading to MetaboAnalyst.
Basic Statistical Analysis is included.
For more information about our different options and levels of metabolome coverage:
At TMIC’s Li Node, we understand that delving into the world of GLOBAL metabolomics profiling can be daunting, especially when considering the potential risks and investments involved in early-stage discovery. To address these challenges and cater to various budgetary needs, we have developed a flexible and staged analysis strategy, focusing on our GLOBAL (Untargeted) Metabolomics by Chemical Isotope Labeling LC-MS.
Our One-channel GLOBAL Metabolomics Analysis is our cost-effective and flexible solution! For just $126 CAD per sample, we offer a basic one-channel analysis. This initial stage allow for the identification of 2000-3000 metabolites (not just features), providing a robust foundation for your research. This approach is not only cost-effective but also incredibly useful in shaping the direction of subsequent, more detailed GLOBAL analysis. The analysis includes:
- Metabolite identification
- Accurate relative concentration measurements
- Basic statistical analysis
How It Works: A Step-by-Step Guide
- Sample Reception: We start by receiving your raw samples, which can include a variety of biological materials such as unprocessed urine, plasma, serum, cell pellets, tissue, etc.
- Aliquoting for Analysis: Each sample is carefully aliquoted, depending on the number of
submetabolomes (channels) required and whether a GLOBAL lipidome analysis is also necessary. - Basic Analysis: Utilizing one aliquot, we conduct the Basic GLOBAL Metabolomics Analysis (one channel) at an affordable rate of $126 CAD per sample.
- Reporting and Consultation: Following the analysis, we provide a detailed report of the findings and engage in a consultation to discuss the results and their implications.
- Client Decision Point:
- Option A: If you decide to pursue further analysis for a more comprehensive coverage, such as Elevated or Comprehensive Metabolomics Analysis and/or lipidome analysis, we seamlessly continue the process. The total cost will be determined based on the number of channels analyzed and the inclusion of lipidome analysis.
- Option B: Should you choose not to proceed with additional analysis, only the cost of the one- channel analysis will be charged.
BASIC GLOBAL Metabolomics by CIL LC-MS
- Academic: $126 CAD/Sample
- Industry: $163.80 CAD/Sample
ELEVATED GLOBAL Metabolomics by CIL LC-MS
- Academic: $224 CAD/sample
- Industry: $291.20 CAD/sample
COMPREHENSIVE GLOBAL Metabolomics by CIL LC-MS:
- Academic: $420 CAD/sample
- Industry: $546 CAD/sample
Flexible GLOBAL Metabolomics by CIL LC-MS:
- Academic:
- $126 CAD/sample for the first channel
- $98 CAD for any additional channel.
For example, choose 3 channels: $126+2x$98=$322 CAD per sample.)
- Industry:
- $163.80 CAD/sample for the first channel
- $127.40 CAD for any additional channel.
For example, choose 3 channels: $163.80+2x$127.40=$418.60 CAD per sample.
The above prices apply to a sample size of 20 or more. For less than 20 samples, an additional $400 CAD applies per channel.
All services require a flat fee of $200 CAD for sample handling per project.
Metabolite identification is based on three-tiered ID approach:
Tier 1: CIL Library based on accurate mass, RT and peak pairs.
Tier 2: Linked Identity Library, high confidence putative identifications (90% accurate) based on accurate mass and predicted RTs.
Tier 3: Putative identification, which is primarily based on mass-to-charge ratio (m/z) data matched to
database hits
Example Data (Human serum; other types of samples should give similar results)
Basic Analysis: a total of 2,051 peak pairs or metabolites are detected and relatively quantified
with high accuracy. Among them, 532 metabolites are identified with high confidence (Tier 1 and Tier 2) that can be used for pathway analysis. 1,313 metabolites are mass-matched (Tier 3). 206 peak pairs are unidentified or unmatched.
Elevated Analysis: a total of 4,196 peak pairs or metabolites are detected and relatively quantified with high accuracy. Among them, 937 metabolites are identified with high confidence (Tier 1 and Tier 2) that can be used for pathway analysis. 2,809 metabolites are mass-matched (Tier 3). 450 peak pairs are unidentified or unmatched.
Comprehensive Analysis: a total of 8,196 peak pairs or metabolites are detected and relatively quantified with high accuracy. Among them, 1,483 metabolites are identified with high confidence (Tier 1 and Tier 2) that can be used for pathway analysis. 5,523 metabolites are mass- matched (Tier 3). 1190 peak pairs are unidentified or unmatched.
Sample Identifications for BASIC GLOBAL Metabolomics by CIL-LC-MS
For the lists of metabolite identifications from the elevated or comprehensive analysis, please Professor Liang Li (liang.li@ualberta.ca)
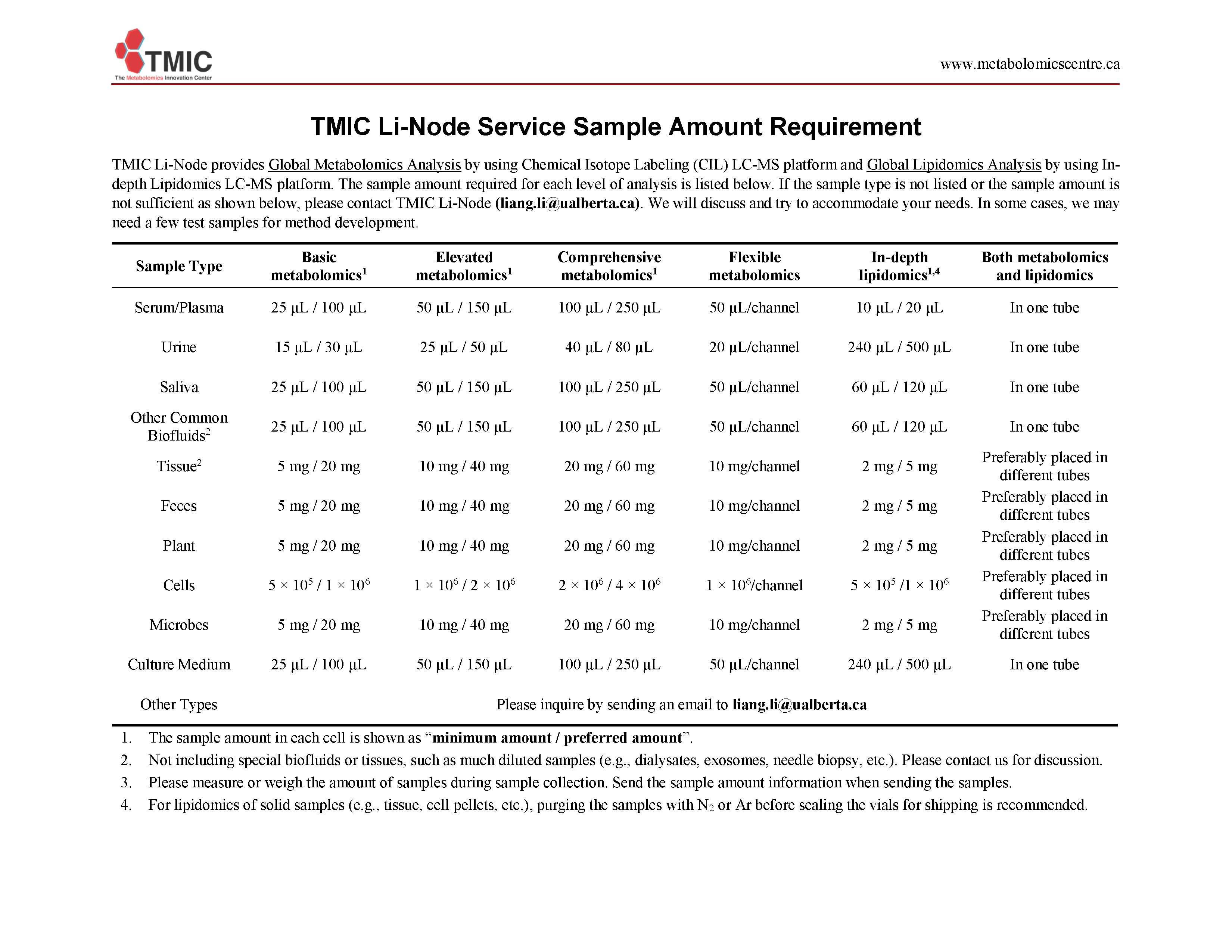
The sample amount required for each level of analysis is listed below. If the sample type is not listed or the sample amount is not sufficient as shown below, please contact TMIC Li-Node (liang.li@ualberta.ca). We will discuss and try to accommodate your needs. In some cases, we may need a few test samples for method development.
Should you require further information or have any inquiries, please visit our Contact Us page.










